PyWGCNA: A python package for weighted gene co-expression network analysis¶
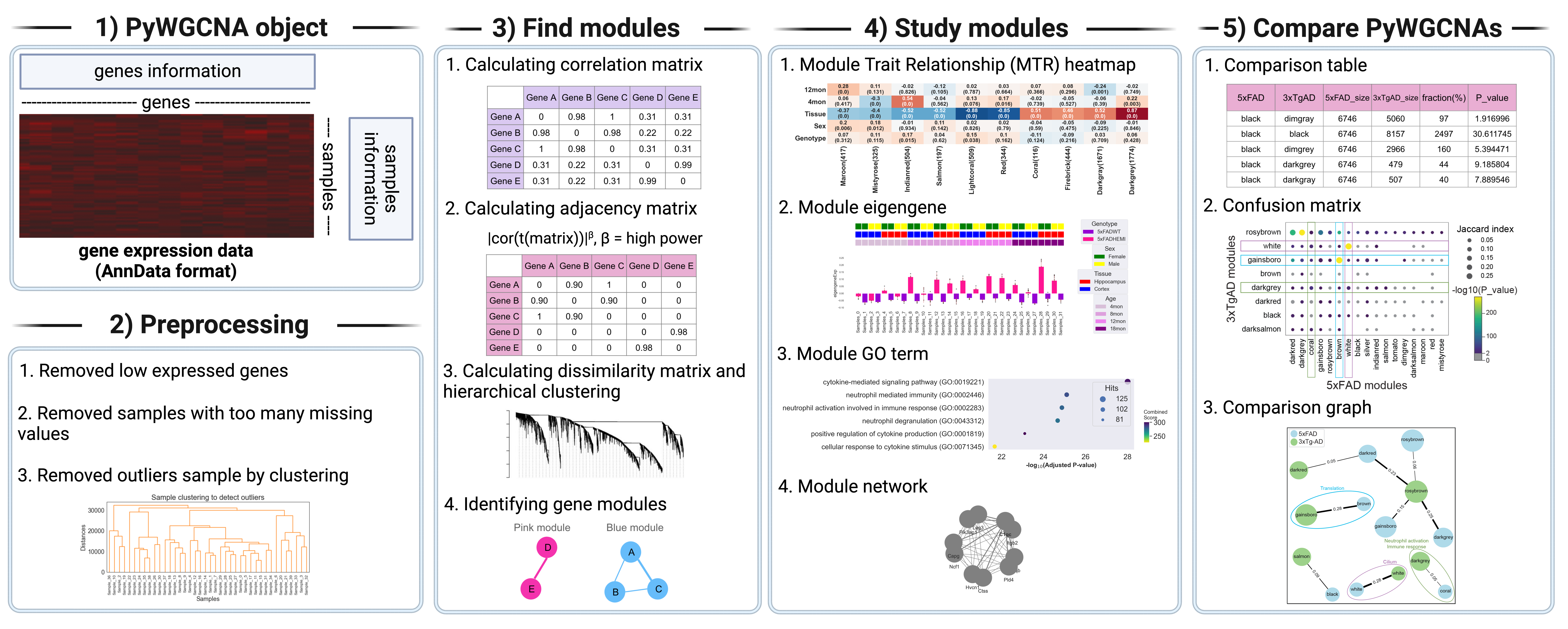
PyWGCNA is a Python library designed to do Weighted correlation network analysis (WGCNA) can be used for finding clusters (modules) of highly correlated genes, for summarizing such clusters using the module eigengene for relating modules to one another and to external sample traits (using eigengene network methodology), and for calculating module membership measures. Users can also compare WGCNA from different datasets including single cell gene markers.
You can find GitHub pages here.